 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 2
Received: 14 Jan., 2024 Accepted: 26 Feb., 2024 Published: 07 Mar., 2024
The livestock sector is under increasing pressure to meet the growing demand for animal products while improving animal health, performance, and reducing environmental impact. Integrative omics approaches, encompassing genomics, transcriptomics, proteomics, epigenomics, and metabolomics, offer promising avenues to address these challenges. These technologies enable a deeper understanding of the genetic and molecular mechanisms underlying economically important traits, such as feed efficiency, meat quality, and disease resistance. Despite significant advancements, the integration of multi-omics data into breeding strategies remains complex, requiring sophisticated analytical methods and better functional genome annotation. Initiatives like the Functional Annotation of Animal Genomes (FAANG) project are pivotal in overcoming these limitations. By leveraging integrative network modeling and multi-tissue transcriptomic profiles, researchers can elucidate the intricate regulatory networks that drive genotype-phenotype associations. This comprehensive approach is expected to enhance the accuracy of genomic predictions and breeding values, ultimately leading to more efficient and sustainable livestock production systems.
1 Introduction
Livestock breeding has long been a cornerstone of agricultural practices, aimed at enhancing desirable traits such as productivity, health, and adaptability in farm animals. Traditional breeding methods have relied heavily on phenotypic selection and quantitative genetics to achieve genetic gains in traits like milk, meat, and egg production. However, these methods often fall short in addressing complex traits such as disease resistance, fertility, and behavior, which are influenced by multiple genetic and environmental factors (Camara et al., 2019; Baes et al., 2022).
As global demand for animal products continues to rise, there is an increasing need for more efficient and sustainable breeding strategies. Modern breeding techniques, such as genomic selection and marker-assisted selection, have revolutionized the field by enabling more precise and accelerated genetic improvements. These methods have been particularly effective in enhancing traits that are difficult to measure or have low heritability (Mutenje et al., 2020). However, the full potential of these techniques is yet to be realized, especially in developing countries where traditional practices still dominate (Camara et al., 2019; Mutenje et al., 2020).
The advent of omics technologies-genomics, transcriptomics, proteomics, epigenomics, and metabolomics-has opened new avenues for understanding the genetic architecture of complex traits in livestock. These technologies allow for a comprehensive analysis of the molecular mechanisms underlying phenotypic variations, thereby facilitating more informed breeding decisions (Diniz and Ward, 2021; Banerjee et al., 2022; Verardo et al., 2023). For instance, integrative network modeling and multi-omics approaches have been employed to untangle the biological mechanisms driving genotype-phenotype associations, thereby improving the accuracy of trait predictions (Diniz and Ward, 2021; Berry et al., 2011). Projects like the Functional Annotation of Animal Genomes (FAANG) have further enriched our understanding by providing extensive datasets on various livestock species (Verardo et al., 2023).
This study aims to explore the integrative use of omics technologies to enhance livestock breeding strategies. By leveraging multi-omics data, we seek to develop more precise and sustainable breeding programs that can meet the growing global demand for animal products. The scope of this study includes a comprehensive review of current omics technologies, their applications in livestock breeding, and the potential challenges and opportunities they present. Through this integrative approach, we hope to provide valuable insights that can guide future research and practical applications in the field of livestock breeding.
2 Genomics in Livestock Breeding
2.1 Introduction to genomics and genomic selection
Genomics has revolutionized livestock breeding by enabling the identification and utilization of genetic markers associated with desirable traits. Traditional breeding methods relied heavily on phenotypic selection, which was often slow and inefficient, especially for traits that are sex-limited or expressed later in life (Chakraborty et al., 2014). Genomic selection (GS) has emerged as a powerful tool, allowing breeders to make selection decisions based on genomic breeding values (GEBV), which are calculated using dense genetic markers spread across the entire genome (Hayes et al., 2009). This approach captures the effects of numerous quantitative trait loci (QTL) that contribute to trait variation, significantly enhancing the accuracy and speed of genetic improvement (Meuwissen et al., 2016).
2.2 Genome-wide association studies (GWAS) and their applications
Genome-Wide Association Studies (GWAS) have been instrumental in identifying genetic variants associated with economically important traits in livestock. By comparing the genetic makeup of animals with different phenotypes, GWAS can pinpoint specific regions of the genome that influence traits such as milk production, growth rate, and disease resistance (Stella et al., 2010). These studies have led to the discovery of numerous QTL and have provided valuable insights into the genetic architecture of complex traits (Diniz and Ward, 2021). For instance, GWAS has been used to identify selection signatures in dairy cattle, revealing genetic differences that have arisen due to selective breeding for milk production (Stella et al., 2010).
2.3 Advances in genomic tools and techniques
The field of genomics has seen significant advancements in tools and techniques, which have further enhanced the effectiveness of genomic selection. The development of high-throughput genotyping technologies has made it possible to genotype animals for hundreds of thousands of single nucleotide polymorphisms (SNPs) in a cost-effective manner (Meuwissen et al., 2016). Additionally, the integration of multi-omics approaches, including transcriptomics, proteomics, and metabolomics, has provided a more comprehensive understanding of the genetic and biological mechanisms underlying complex traits (Diniz and Ward, 2021; Eenennaam et al., 2014). These advancements have improved the accuracy of genomic predictions and have facilitated the development of more effective breeding strategies (Chakraborty et al., 2022).
2.4 Case study: genomic selection in cattle breeding
Genomic selection has had a profound impact on cattle breeding, particularly in the dairy industry. By using GEBV, breeders can select young bulls with high genetic potential without waiting for progeny test results, thereby reducing the generation interval and accelerating genetic gain (Hayes et al., 2009). Studies have shown that the reliability of GEBV for young bulls can range from 20% to 67%, depending on factors such as the heritability of the trait and the size of the reference population (Hayes et al., 2009). The implementation of GS in dairy cattle has already led to significant improvements in milk production and other economically important traits (Verardo et al., 2023). Moreover, the use of whole-genome sequence data is anticipated to further increase the accuracy of GS by including causative mutations in the data. This case study highlights the transformative potential of genomic selection in livestock breeding and underscores the importance of continued research and development in this field (Meuwissen et al., 2016; Hayes et al., 2009; Verardo et al., 2023).
3 Transcriptomics: Unveiling Gene Expression Patterns
3.1 Basics of transcriptomics in livestock
Transcriptomics involves the study of RNA transcripts produced by the genome under specific circumstances or in a specific cell. In livestock, transcriptomics is essential for understanding the molecular mechanisms underlying various traits, including growth, reproduction, and disease resistance. By analyzing RNA sequences, researchers can identify which genes are active, how their expression levels change in different tissues, and how these changes correlate with phenotypic traits (Fang et al., 2020; Arishima et al., 2022).
3.2 Techniques for transcriptomic analysis
Several techniques are employed in transcriptomic analysis, with RNA sequencing (RNA-seq) being the most prominent. RNA-seq allows for the comprehensive profiling of all transcripts in a sample, providing insights into gene expression levels, alternative splicing events, and novel transcript discovery. High-throughput sequencing technologies, such as short-read and long-read sequencing, are commonly used to capture the complexity of the transcriptome (Foissac et al., 2019; Arishima et al., 2022). Additionally, integrative approaches combining transcriptomics with other omics data, such as proteomics and epigenomics, enhance the understanding of gene regulation and functional genomics (Kumar et al., 2016; Qin et al., 2016).
3.3 Applications of transcriptomics in breeding programs
Transcriptomics has numerous applications in livestock breeding programs. By identifying genes associated with economically important traits, such as milk production, meat quality, and disease resistance, transcriptomic data can inform selective breeding strategies. For instance, integrative analyses of tissue-specific genes with genome-wide association studies (GWAS) have identified candidate genes and relevant tissues for traits like male fertility and body conformation in cattle (Fang et al., 2020). Moreover, transcriptomic data can be used to improve genome annotation, predict gene function, and develop genomic selection models that enhance breeding efficiency (Verardo et al., 2023; Diniz and Ward, 2021).
3.4 Case study: transcriptomic insights into disease resistance
A comprehensive analysis of 124 transcriptomes from various tissues in Japanese Black cattle revealed significant insights into disease resistance. By examining the expression profiles of causative genes for genetic disorders, researchers identified disease-relevant expression patterns that could be targeted in breeding programs to enhance disease resistance (Figure 1) (Arishima et al., 2022). Another study involving 723 RNA-seq data from cattle tissues identified tissue-specific genes and their roles in immune response, providing a valuable resource for understanding the genetic basis of disease resistance and developing strategies to improve livestock health (Fang et al., 2020). These findings underscore the potential of transcriptomics to uncover the molecular mechanisms underlying disease resistance and inform breeding strategies aimed at producing healthier livestock.
 Figure 1 Japanese Black cattle and Wagyu Genome Database (Adopted from Arishima et al., 2022) Image caoption: This figure appears to illustrate the phenotypic (physical) characteristics of Japanese Black cattle and links them with genetic data available in a specialized genome database. The Wagyu Genome Database section (Part B) emphasizes the available genetic resources for research, particularly in understanding the genomic factors that contribute to the cattle's desirable traits, such as high-quality meat production. This database would be a useful tool for researchers focusing on genetic studies, breeding, and enhancing the traits of Wagyu cattle (Adopted from Arishima et al., 2022) |
4 Proteomics: Understanding Protein Dynamics
4.1 Role of proteomics in livestock breeding
Proteomics, the large-scale study of proteins, plays a crucial role in livestock breeding by providing insights into the molecular mechanisms underlying various traits. Proteomics allows researchers to monitor in vivo performances of livestock animals, such as growth, fertility, and milk quality, and to understand the molecular processes that affect meat quality (D’Alessandro and Zolla, 2013). By identifying and validating biomarkers associated with important traits, proteomics can enhance the selection process in breeding programs, leading to improved animal performance and productivity (Long, 2020).
4.2 Techniques for protein profiling
Several advanced techniques are employed in proteomic analysis to study protein composition, structure, function, and interactions. Common methods include 2D gel electrophoresis, MALDI-TOF/MS, X-ray crystallography, NMR, protein microarrays, two-hybrid screening, and western blotting (Mote and Filipov, 2020). These techniques enable the generation of large proteomic datasets, which can be used to identify changes in protein expression, interactions, or modifications. The integration of these datasets with other omics data provides a comprehensive view of cellular functions and their regulation (Loor et al., 2015; Dihazi et al., 2018).
4.3 Application of proteomics in trait selection
Proteomics has been extensively applied to understand and improve various traits in livestock. For instance, changes in protein profiles during myogenesis have been studied in cattle, pigs, and fowl, providing insights into key stages of muscle development and identifying processes that are similar or divergent between species (Picard et al., 2010). Proteomic studies have also identified biological markers of meat quality, such as tenderness, and have helped to understand the biochemical mechanisms underlying the effects of stress during the pre-slaughter period on meat quality traits (Picard et al., 2010). These findings can be used to select animals with desirable traits, thereby enhancing breeding strategies.
4.4 Case study: proteomic approaches in enhancing meat quality
A notable application of proteomics in livestock breeding is the enhancement of meat quality. Proteomic studies have revealed that meat quality is influenced by a complex interplay of genetic and extrinsic factors, such as rearing environment, feeding conditions, physical activity, and pre-slaughter handling (D’Alessandro and Zolla, 2013). For example, post-mortem alterations to the muscle proteome reflect the biological complexity of the "muscle to meat conversion" process, which is crucial for determining meat tenderness (D’Alessandro and Zolla, 2013). By integrating proteomic data with other omics approaches, researchers have established milestones in understanding the events leading to meat quality, enabling the development of strategies to produce high-quality meat (Picard et al., 2010; D’Alessandro and Zolla, 2013).
5 Metabolomics: Profiling Biochemical Pathways
5.1 Introduction to metabolomics in livestock
Metabolomics is a powerful omics technology that involves the comprehensive analysis of small molecule metabolites within biological samples such as cells, tissues, and biofluids. This approach provides a detailed snapshot of the metabolic state of an organism, reflecting the end-products of complex genetic, epigenetic, and environmental interactions (Goldansaz et al., 2017). In livestock research, metabolomics has been increasingly utilized to enhance phenotypic characterization, offering insights into animal health, disease diagnosis, and economically important traits such as feed efficiency and milk production (Fontanesi, 2016; Goldansaz et al., 2017). The integration of metabolomics with other omics data, such as genomics, has the potential to refine trait descriptions and improve the prediction of breeding values, thereby advancing livestock breeding strategies (Fontanesi, 2016).
5.2 Techniques for metabolic profiling
Metabolomics employs advanced analytical techniques, primarily mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy, to identify and quantify metabolites. High-resolution mass spectrometry, coupled with gas or liquid chromatography, is particularly effective for the accurate measurement of a wide range of metabolites. Techniques such as reversed-phase liquid chromatography (RPLC) and hydrophilic interaction liquid chromatography (HILIC) are used to separate lipophilic and hydrophilic metabolites, respectively, allowing for comprehensive coverage of the metabolome. Recent advancements in integrative multi-omics approaches have enabled the simultaneous analysis of proteins and metabolites from a single sample, providing a more holistic view of the biological system (Blum et al., 2018).
5.3 Impact of metabolomics on breeding decisions
The application of metabolomics in livestock breeding can significantly impact breeding decisions by providing detailed phenotypic data that complements genomic information. Metabolomics can identify biomarkers associated with desirable traits, such as growth rate, milk production, and fat deposition, which can be used to select animals with superior genetic potential (Fontanesi, 2016). Additionally, metabolomics can help elucidate the biochemical pathways underlying these traits, offering insights into the metabolic mechanisms that drive phenotypic variation (Fontanesi, 2016; Goldansaz et al., 2017). By integrating metabolomic data with genomic and other omics data, researchers can develop more accurate models for predicting breeding values and optimizing selection programs.
5.4 Case study: metabolomics in nutritional efficiency
A notable application of metabolomics in livestock research is the study of nutritional efficiency. Metabolomic profiling can reveal how different diets affect the metabolic pathways in livestock, identifying key metabolites and pathways associated with improved feed efficiency (Goldansaz et al., 2017). For example, a study on cattle metabolomics identified specific metabolites linked to better feed conversion ratios, which could be used as biomarkers for selecting animals with higher nutritional efficiency (Goldansaz et al., 2017). This approach not only enhances the understanding of the metabolic basis of feed efficiency but also provides practical tools for improving livestock production through targeted breeding strategies. Integrative analysis of metabolomics data with other omics layers, such as genomics and transcriptomics, further enhances the ability to identify genetic markers associated with nutritional efficiency, paving the way for more efficient and sustainable livestock production systems (Figure 2) (Jendoubi, 2021).
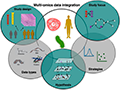 Figure 2 Conceptual overview of multi-omics data integration in the context of biological research (Adopted from: Jendoubi, 2021) Image caption: The figure is divided into several overlapping sections, each representing a key component of a multi-omics approach, showing how these components intersect to create a comprehensive research framework (Adopted from: Jendoubi, 2021) |
6 Integrative Omics Approaches
6.1 Concept and importance of multi-omics integration
The concept of multi-omics integration involves the comprehensive analysis of various omics data types, such as genomics, transcriptomics, proteomics, and metabolomics, to gain a holistic understanding of biological systems. This integrative approach is crucial for elucidating the complex interactions and regulatory mechanisms that underlie phenotypic traits. By combining data from different omics layers, researchers can uncover the intricate networks and pathways that drive biological processes, leading to more accurate predictions and better-informed breeding strategies (Suravajhala et al., 2016; Subramanian et al., 2020; Yang et al., 2021).
6.2 Strategies for integrating genomic, transcriptomic, proteomic, and metabolomic data
Several strategies have been developed to integrate multi-omics data effectively. These include data-driven approaches, which rely on statistical and computational methods to identify correlations and interactions between different omics layers, and knowledge-based approaches, which use existing biological knowledge to guide the integration process. Simultaneous and step-wise integration methods are also employed to combine data from multiple omics layers in a coherent manner. Tools such as mixOmics for R software have been specifically designed to address data integration issues, enabling researchers to perform comprehensive analyses and derive meaningful insights from heterogeneous datasets (Duruflé et al., 2020; Subramanian et al., 2020; Wörheide et al., 2021).
6.3 Benefits and challenges of integrative omics in breeding
The integration of multi-omics data offers numerous benefits for livestock breeding. It enhances the understanding of the genetic architecture underlying important economic traits, improves the accuracy of genomic predictions, and facilitates the identification of biomarkers for disease resistance and performance traits. However, several challenges remain, including the high dimensionality and heterogeneity of omics data, the need for robust bioinformatics tools, and the complexity of modeling interactions between different biological layers. Despite these challenges, initiatives such as the Functional Annotation of Animal Genomes (FAANG) project are making significant strides in addressing these issues and advancing the field of integrative omics (Figure 3) (Suravajhala et al., 2016; Diniz and Ward, 2021; Verardo et al., 2023).
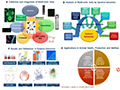 Figure 3 Overview of integrated genomics with various other ‘omics’ platforms/data types created via array-based or spectrometry or NGS technologies and systems genomics analyses (Adopted from Suravajhala et al., 2016) Image caption: This figure is an overview of integrated genomics with various ‘omics’ platforms/data types and systems genomics analyses, illustrating how different types of biological data can be collected, analyzed, validated, and applied in animal health, production, and welfare (Adopted from Suravajhala et al., 2016) |
6.4 Case study: multi-omics approach to improve dairy production
A notable example of the application of multi-omics integration in livestock breeding is the improvement of dairy production. By combining genomic, transcriptomic, proteomic, and metabolomic data, researchers have been able to identify key regulatory mechanisms and genetic variants associated with milk yield and quality. For instance, multi-tissue transcriptomic profiling has revealed differential tissue regulation mechanisms in nutrient-restricted bovine fetuses, providing insights into the genetic basis of milk production traits. These findings have the potential to inform breeding programs aimed at enhancing dairy production efficiency and sustainability (Diniz and Ward, 2021; Pazhamala et al., 2021; Verardo et al., 2023).
7 Case Study: Integrative Omics Application in Livestock Breeding
7.1 Background of the selected case study
The selected case study focuses on the application of integrative omics approaches in cattle breeding. Traditional quantitative genetics has significantly advanced the selection of cattle for specific traits, yet considerable phenotypic variation remains unexplained. This gap represents an opportunity for further improvement in animal production. The advent of omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, has revolutionized the understanding of the genetic architecture controlling traits of interest. These technologies offer a systems biology approach to animal breeding, providing a comprehensive understanding of the biological mechanisms underlying phenotypic traits (Berry et al., 2011; Diniz and Ward, 2021).
7.2 Omics data integration process
The integration of omics data involves combining information from various biological layers to create a holistic view of the genetic and phenotypic landscape. In cattle breeding, this process typically starts with the collection of high-throughput data from genomics, transcriptomics, proteomics, and metabolomics. Bioinformatics tools are then employed to analyze these datasets, identifying key genes, proteins, and metabolites involved in important traits. For instance, multi-tissue transcriptomic profiles can be used to uncover mechanisms driving differential tissue regulation under specific conditions, such as nutrient restriction (Diniz and Ward, 2021). The integration process also includes the use of network modeling to understand the interactions between different biological layers and their impact on phenotypic traits (Loor et al., 2015).
7.3 Key findings and impact on breeding strategies
The application of integrative omics in cattle breeding has led to several key findings. For example, studies have identified quantitative trait loci (QTL) and causal genes associated with traits like feed efficiency and meat quality. Integrative approaches have also revealed the significant role of the gut microbiome in shaping animal performance, highlighting the complex interplay between the host genome and its microbiota (Diniz and Ward, 2021). These findings have profound implications for breeding strategies, enabling more accurate genomic predictions and the development of biology-driven breeding programs. By incorporating omics data into breeding equations, researchers can improve the prediction accuracy of economically important traits, ultimately enhancing production efficiency and sustainability (Mote and Filipov, 2020; Verardo et al., 2023).
7.4 Lessons learned and future implications
The integration of omics data in livestock breeding has provided valuable lessons. One major lesson is the importance of a well-annotated genome to fully understand the genetic basis of complex traits. Despite advances in network modeling, there is still a need for better functional annotation of genomes to bridge the gap between genotype and phenotype (Diniz and Ward, 2021). Additionally, the integration of multi-omics data poses significant analytical challenges, requiring the development of robust statistical models and bioinformatics tools. Future research should focus on addressing these challenges and exploring the potential of integrative omics in other livestock species and traits. The continued advancement of omics technologies and their application in breeding programs will be crucial for meeting the growing demand for animal products while ensuring sustainability and animal welfare (Berry et al., 2011; Loor et al., 2015; Ribeiro et al., 2020).
8 Ethical, Legal, and Social Implications (ELSI) of Omics Technologies
8.1 Ethical considerations in omics-based breeding
The application of omics technologies in livestock breeding raises several ethical concerns. One primary issue is the welfare of the animals involved. The intensive selection for specific traits, such as increased productivity or disease resistance, may inadvertently lead to negative health outcomes or reduced genetic diversity, which can compromise animal welfare (Diniz and Ward, 2021; Berry et al., 2011). Additionally, the manipulation of genetic material through techniques such as genome editing poses ethical questions about the extent to which humans should interfere with natural genetic processes. The potential for unintended consequences, such as off-target effects or the creation of new diseases, necessitates careful consideration and regulation (Chakraborty et al., 2022). Furthermore, the transparency and consent in the use of genetic data from animals are crucial. Farmers and breeders must be fully informed about the implications of using omics technologies and the potential risks and benefits involved.
8.2 Legal and regulatory aspects
The legal landscape surrounding the use of omics technologies in livestock breeding is complex and varies significantly across different regions. Regulatory frameworks must balance the promotion of innovation with the protection of animal welfare and public health. For instance, the use of genome editing technologies, such as CRISPR, is subject to stringent regulations in many countries to ensure that any modifications are safe and ethically justified. Additionally, there are intellectual property considerations, as the development of new breeds or genetic lines using omics technologies can lead to patenting and ownership issues. This can impact the accessibility and affordability of these technologies for small-scale farmers and breeders (Verardo et al., 2023; Yang et al., 2021). International cooperation and harmonization of regulations are essential to facilitate the safe and ethical use of omics technologies in livestock breeding.
8.3 Social and economic implications for farmers and industry
The integration of omics technologies into livestock breeding has significant social and economic implications. On one hand, these technologies can lead to substantial improvements in productivity, disease resistance, and overall animal health, which can enhance the profitability and sustainability of livestock farming (Berry et al., 2011; Chakraborty et al., 2022). However, the high cost of omics technologies and the need for specialized knowledge and infrastructure can be prohibitive for small-scale farmers, potentially widening the gap between large commercial operations and smaller farms (Verardo et al., 2023). Additionally, the adoption of these technologies may lead to changes in traditional farming practices and rural livelihoods, as farmers may need to adapt to new breeding strategies and management practices (Mote and Filipov, 2020; Subramanian et al., 2020). It is crucial to ensure that the benefits of omics technologies are equitably distributed and that support is provided to farmers to facilitate their adoption and integration into existing systems. This includes providing education, training, and financial assistance to help farmers navigate the transition to omics-based breeding practices.
9 Future Directions and Innovations
9.1 Emerging trends in omics technologies
The rapid advancement of omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, is revolutionizing livestock breeding strategies. These technologies enable the comprehensive analysis of genetic and phenotypic data, facilitating the identification of genes and pathways responsible for economically important traits (Mahmood et al., 2022). High-throughput sequencing and integrative network modeling are particularly promising, as they allow for the detailed mapping of complex traits and the interactions between different regulatory layers (Yang et al., 2017). The integration of multi-omics data with machine learning algorithms is expected to further enhance the precision and efficiency of breeding programs, enabling the development of livestock that are more resilient to environmental stresses and diseases (Verardo et al., 2023).
9.2 Potential for personalized breeding programs
The concept of personalized breeding programs is gaining traction, driven by the ability to analyze multi-omics data at the individual level. This approach allows for the customization of breeding strategies to optimize the genetic potential of each animal, taking into account its unique genetic makeup and environmental interactions. Precision breeding, which combines genomic selection with genome editing techniques, is expected to become a crucial practice in future livestock breeding. This method not only improves the accuracy of breeding value estimation but also enables the selection of genetically superior and disease-free animals at an early stage of life, thereby enhancing productivity and profitability (Kaur et al., 2021).
10 Concluding Remarks
The integration of various omics technologies has revolutionized livestock breeding strategies by providing a comprehensive understanding of the genetic and molecular bases of economically important traits. Genomics, transcriptomics, proteomics, metabolomics, and epigenomics have all contributed to this advancement. These technologies have enabled the identification of quantitative trait loci (QTL) and causal genes, although the number of identified causal mutations remains limited. The integration of multi-omics data has facilitated the development of systems biology approaches, which allow for the modeling of complex biological networks and the prediction of phenotypic outcomes. For instance, combining multi-tissue transcriptomic profiles has helped identify mechanisms driving tissue-specific regulation in nutrient-restricted bovine fetuses. Additionally, the Functional Annotation of Animal Genomes (FAANG) project has been instrumental in generating datasets to decipher genome functions across various livestock species.
Future research should focus on further integrating multi-omics data to enhance the accuracy of genomic predictions and breeding values. This includes the development of advanced statistical models and bioinformatics tools capable of handling and interpreting large-scale omics datasets. There is also a need to improve the functional annotation of livestock genomes to better understand the genetic basis of complex traits. Moreover, exploring the interactions between the host genome and gut microbiota can provide new insights into animal performance and health.
In practice, the adoption of precision breeding techniques that utilize multi-omics information should be encouraged. This approach can lead to more accurate and personalized breeding strategies, ultimately improving production efficiency and sustainability. Additionally, integrating omics data with traditional breeding methods can help fine-tune nutritional management and other husbandry practices to optimize animal health and productivity. Finally, continued support for initiatives like FAANG and the development of open-access databases will be crucial for advancing livestock breeding research and translating scientific discoveries into practical applications.
Acknowledgements
Author would like to express our gratitude to the two anonymous peer reviewers for their critical assessment and constructive suggestions on our manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Arishima T., Wakaguri H., Nakashima R., Sakakihara S., Kawashima K., Sugimoto Y., Suzuki Y., and Sasaki S., 2022, Comprehensive analysis of 124 transcriptomes from 31 tissues in developing, juvenile, and adult Japanese Black cattle. DNA Research, 29(5):dsac032.
https://doi.org/10.1093/dnares/dsac032
PMID: 36047829 PMCID: PMC9555877
Baes C.F., Rochus C.M., Houlahan K., Jr G.A., van Staaveren N., and Miglior F., 2022, 22 sustainable livestock breeding: challenges and opportunities, Journal of Animal Science, 100(Suppl 3): 13.
https://doi.org/10.1093/jas/skac247.023
Banerjee P., Cesar A.S.M., and De Lima A.O., 2022, Editorial: gene regulation explored by systems biology in livestock science, Frontiers in Genetics, 13: 859061.
https://doi.org/10.3389/fgene.2022.859061
PMID: 35464851 PMCID: PMC9027333
Benedetto A., Pezzolato M., Biasibetti E., and Bozzetta E., 2021, Omics applications in the fight against abuse of anabolic substances in cattle: challenges, perspectives and opportunities, Current Opinion in Food Science, 40: 112-120.
https://doi.org/10.1016/J.COFS.2021.03.001
Berry D.P., Meade K.G., Mullen M.P., Butler S., Diskin M.G., Morris D., and Creevey C.J., 2011, The integration of 'omic' disciplines and systems biology in cattle breeding, Animal : an international journal of animal bioscience, 5(4): 493-505.
https://doi.org/10.1017/S1751731110002120
PMID: 22439945
Bhattacharya A., Li Y., and Love M.I., 2020, MOSTWAS: multi-omic strategies for transcriptome-wide association studies, PLoS Genetics, 17(3): e1009398.
https://doi.org/10.1101/2020.04.17.047225
PMID: 33684137 PMCID: PMC7971899
Blum B.C., Mousavi F., and Emili A., 2018, Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis, Molecular omics, 14(5): 307-319.
https://doi.org/10.1039/c8mo00136g
PMID: 30211418
Camara Y., Moula N., Sow F., Sissokho M.M., and Antoine-Moussiaux N., 2019, Analysing innovations among cattle smallholders to evaluate the adequacy of breeding programs, Animal : an international journal of animal bioscience, 13(2): 417-426.
https://doi.org/10.1017/S1751731118001544
Chakraborty D., Sharma N., Kour S., Sodhi S.S., Gupta M.K., Lee S.J., and Son Y.O., 2022, Applications of omics technology for livestock selection and improvement, Frontiers in Genetics, 13: 774113.
https://doi.org/10.3389/fgene.2022.774113
PMID: 35719396 PMCID: PMC9204716
D’Alessandro A., and Zolla L., 2013, Meat science: from proteomics to integrated omics towards system biology, Journal of Proteomics, 78: 558-577.
https://doi.org/10.1016/j.jprot.2012.10.023
PMID: 23137709
Dihazi H., Asif A.R., Beissbarth T., Bohrer R., Feussner K., Feussner I., Jahn O., Lenz C., Majcherczyk A., Schmidt B., Schmitt K., Urlaub H., and Valerius O., 2018, Integrative omics - from data to biology, Expert Review of Proteomics, 15: 463-466.
https://doi.org/10.1080/14789450.2018.1476143
PMID: 29757692
Diniz W., and Ward A., 2021, 282 Multi-omics approaches to improve animal production, Journal of Animal Science, 99: 20-21.
https://doi.org/10.1093/JAS/SKAB054.036
Duruflé H., Selmani M., Ranocha P., Jamet E., Dunand C., and Déjean S., 2020, A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis, Briefings in Bioinformatics, 22(3): bbaa166.
https://doi.org/10.1093/bib/bbaa166
PMID: 32778869
Eenennaam A.L.V., Weigel K.A., Young A.E., Cleveland M., and Dekkers J., 2014, Applied animal genomics: results from the field, Annual Review of Animal Biosciences, 2: 105-139.
https://doi.org/10.1146/annurev-animal-022513-114119
PMID: 25384137
Fang L.Z., Cai W.T., Liu S.L., Canela‐Xandri O., Gao Y.H., Jiang J., Rawlik K., Li B., Schroeder S., Rosen B., Li C., Sonstegard T., Alexander L., Tassell C., VanRaden P., Cole J.B., Yu Y., Zhang S.L., Tenesa A., Ma L., and Liu G.E., 2020, Comprehensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle, Genome Research, 30(5): 790-801.
https://doi.org/10.1101/gr.250704.119
PMID: 32424068 PMCID: PMC7263193
Foissac S., Djebali S., Munyard K., Vialaneix N., Rau A., Acloque H., Lagarrigue S., and Giuffra E., 2019, 32 Functional annotation of livestock genomes: chromatin structure and regulation of gene expression, Journal of Animal Science, 97(Suppl 2): 15-16.
https://doi.org/10.1093/JAS/SKZ122.028
Fontanesi L., 2016, Metabolomics and livestock genomics: Insights into a phenotyping frontier and its applications in animal breeding, Animal Frontiers, 6(1): 73-79.
https://doi.org/10.2527/AF.2016-0011
Goldansaz S.A., Guo A.C., Sajed T., Steele M., Plastow G., and Wishart D., 2017, Livestock metabolomics and the livestock metabolome: a systematic review, PLoS One, 12(5): e0177675.
https://doi.org/10.1371/journal.pone.0177675
PMID: 28531195 PMCID: PMC5439675
Hayes B.J., Bowman P.J., Chamberlain A., and Goddard M., 2009, Invited review: genomic selection in dairy cattle: progress and challenges, Journal of Dairy Science, 92(2) 433-443.
https://doi.org/10.3168/jds.2008-1646
PMID: 19164653
Ivanisevic J., Zhu Z.J., Plate L., Tautenhahn R., Chen S., O'Brien P.J., Johnson C.H., Marletta M., Patti G.J., and Siuzdak G., 2013, Toward 'omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism, Analytical Chemistry, 85(14): 6876-6884.
https://doi.org/10.1021/ac401140h
PMID: 23781873 PMCID: PMC3761963
Jendoubi T., 2021, Approaches to integrating metabolomics and multi-omics data: a primer, Metabolites, 11(3): 184.
https://doi.org/10.3390/metabo11030184
PMID: 33801081 PMCID: PMC8003953
Jendoubi T., and Ebbels T., 2018, Integrative analysis of time course metabolic data and biomarker discovery, BMC Bioinformatics, 21(1): 11.
https://doi.org/10.1186/s12859-019-3333-0
PMID: 31918658 PMCID: PMC6953149
Jonas E., and de Koning D.J., 2015, Genomic selection needs to be carefully assessed to meet specific requirements in livestock breeding programs, Frontiers in Genetics, 6: 49.
https://doi.org/10.3389/fgene.2015.00049
PMID: 25750652 PMCID: PMC4335173
Kaur B., Sandhu K.S., Kamal R., Kaur K., Singh J., Röder M.S., and Muqaddasi Q.H., 2021, Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: applications, challenges, and prospects, Plants, 10(10): 1989.
https://doi.org/10.3390/plants10101989
PMID: 34685799 PMCID: PMC8541486
Kumar D., Bansal G., Narang A., Basak T., Abbas T., and Dash D., 2016, Integrating transcriptome and proteome profiling: Strategies and applications, Proteomics, 16(19): 2533-2544.
https://doi.org/10.1002/pmic.201600140
Langridge P., and Fleury D., 2011, Making the most of 'omics' for crop breeding, Trends in Biotechnology, 29(1): 33-40.
https://doi.org/10.1016/j.tibtech.2010.09.006
PMID: 21030098
Li S.S., Wang Q.J., Lin X., Jin X., Liu L., Wang C., Chen Q., Liu J.X., and Liu H.Y., 2017, The use of “omics” in lactation research in dairy cows, International Journal of Molecular Sciences, 18(5): 983.
https://doi.org/10.3390/ijms18050983
PMID: 28475129 PMCID: PMC5454896
Long J.A., 2020, The 'omics' revolution: Use of genomic, transcriptomic, proteomic and metabolomic tools to predict male reproductive traits that impact fertility in livestock and poultry, Animal Reproduction Science, 220: 106354.
https://doi.org/10.1016/j.anireprosci.2020.106354
PMID: 32482486
Loor J.J., Bionaz M., and Drackley J.K., 2013, Systems physiology in dairy cattle: nutritional genomics and beyond, Annual Review of Animal Biosciences, 1: 365-92
https://doi.org/10.1146/annurev-animal-031412-103728
PMID: 25387024
Ma A.J., McDermaid A., Xu J.Z., Chang Y.Z., and Ma Q., 2020, Integrative methods and practical challenges for single-cell multi-omics, Trends in Biotechnology, 38(9): 1007-1022.
https://doi.org/10.1016/j.tibtech.2020.02.013
PMID: 32818441 PMCID: PMC7442857
Mahmood U., Li X.D., Fan Y.H., Chang W., Niu Y., Li J.N., Qu C.M., and Lu K., 2022, Multi-omics revolution to promote plant breeding efficiency, Frontiers in Plant Science, 13: 1062952.
https://doi.org/10.3389/fpls.2022.1062952
PMID: 36570904 PMCID: PMC9773847
Meuwissen T., Hayes B., and Goddard M., 2013, Accelerating improvement of livestock with genomic selection, Annual Review of Animal Biosciences, 1: 221-237
https://doi.org/10.1146/annurev-animal-031412-103705
PMID: 25387018
Meuwissen T., Hayes B., and Goddard M., 2016, Genomic selection: a paradigm shift in animal breeding, Animal Frontiers, 6(1): 6-14.
https://doi.org/10.2527/AF.2016-0002
Mote R.S., and Filipov N.M., 2020, Use of integrative interactomics for improvement of farm animal health and welfare: an example with fescue toxicosis, Toxins, 12(10): 633.
https://doi.org/10.3390/toxins12100633
PMCID: PMC7600642 PMID: 33019560
Mutenje M., Chipfupa U., Mupangwa W., Nyagumbo I., Manyawu G., Chakoma I., and Gwiriri L., 2020, Understanding breeding preferences among small-scale cattle producers: implications for livestock improvement programmes, Animal, 14(8): 1757-1767.
https://doi.org/10.1017/S1751731120000592
PMID: 32252847
Pazhamala L.T., Kudapa H., Weckwerth W., Millar A.H., and Varshney R.K., 2021, Systems biology for crop improvement, The Plant Genome, 14(2): e20098.
https://doi.org/10.1002/tpg2.20098
PMID: 33949787
Picard B., Berri C., Lefaucheur L., Molette C., Sayd T., and Terlouw C., 2010, Skeletal muscle proteomics in livestock production, Briefings in Functional Genomics, 9(3): 259-278.
https://doi.org/10.1093/bfgp/elq005
PMID: 20308039
Qin J., Yan B., Hu Y.H., Wang P.W., and Wang J.W., 2016, Applications of integrative OMICs approaches to gene regulation studies, Quantitative Biology, 4: 283-301.
https://doi.org/10.1007/s40484-016-0085-y
Raza A., 2020, Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants, Plant Cell Reports, 41(3): 741-763.
https://doi.org/10.1007/s00299-020-02635-8
Ribeiro D.M., Salama A., Vítor A., Arguello A., Moncau C., Santos E., Caja G., Oliveira J., Balieiro J., Hernández-Castellano L., Zachut M., Poleti M., Castro N., Alves S., and Almeida A., 2020, The application of omics in ruminant production: a review in the tropical and sub-tropical animal production context, Journal of Proteomics, 227: 103905.
https://doi.org/10.1016/j.jprot.2020.103905
Stanberry L., Mias G.I., Haynes W., Higdon R., Snyder M., and Kolker E., 2013, Integrative analysis of longitudinal metabolomics data from a personal multi-omics profile, Metabolites, 3(3): 741-760.
https://doi.org/10.3390/metabo3030741
PMID: 24958148 PMCID: PMC3901289
Stella A., Ajmone-Marsan P., Lazzari B., and Boettcher P., 2010, Identification of selection signatures in cattle breeds selected for dairy production, Genetics, 185(4): 1451-1461.
https://doi.org/10.1534/genetics.110.116111
PMID: 20479146 PMCID: PMC2927769
Subramanian I., Verma S., Kumar S., Jere A., and Anamika K., 2020, Multi-omics data integration, interpretation, and its application, Bioinformatics and Biology Insights, 14: 1177932219899051.
https://doi.org/10.1177/1177932219899051
PMID: 32076369 PMCID: PMC7003173
Suravajhala P., Kogelman L.J.A., and Kadarmideen H.N., 2016, Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare, Genetics Selection Evolution, 48(1): 38.
https://doi.org/10.1186/s12711-016-0217-x
PMID: 27130220 PMCID: PMC4850674
Verardo L.L., Brito L.F., Carolino N., and Magalhães A.F.B., 2023, Editorial: omics applied to livestock genetics, Frontiers in Genetics, 14: 1155611.
https://doi.org/10.3389/fgene.2023.1155611
PMID: 36873944 PMCID: PMC9978907
Weckwerth W., 2003, Metabolomics in systems biology, Annual Review of Plant Biology, 54: 669-689.
https://doi.org/10.1146/ANNUREV.ARPLANT.54.031902.135014
PMID: 14503007
Wörheide M.A., Krumsiek J., Kastenmüller G., and Arnold M., 2021, Multi-omics integration in biomedical research-a metabolomics-centric review, Analytica Chimica Acta, 1141: 144-162.
https://doi.org/10.1016/j.aca.2020.10.038
PMID: 33248648 PMCID: PMC7701361
Yang Y.D., Saand M.A., Huang L.Y., Abdelaal W.B., Zhang J., Wu Y., Li J., Sirohi M.H., and Wang F.Y., 2021, Applications of multi-omics technologies for crop improvement, Frontiers in Plant Science, 12: 563953.
https://doi.org/10.3389/fpls.2021.563953
PMID: 34539683 PMCID: PMC8446515
Yang Y.L., Zhou R., and Kui L., 2017, Future livestock breeding: Precision breeding based on multi-omics information and population personalization, Journal of Integrative Agriculture, 16: 2784-2791.
https://doi.org/10.1016/S2095-3119(17)61780-5
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Haimei Wang
Related articles
. Integrative omics
. Livestock breeding
. Genomic selection
. Multi-omics data
. Functional genome annotation
Tools
. Post a comment


